品质至上,客户至上,您的满意就是我们的目标
当前位置: 首页 > 新闻动态
科学家利用Videometer多光谱成像结合多组学技术揭示紫花苜蓿硬实种子的综合休眠模式
发表时间: 点击:1135
来源:北京博普特科技有限公司
分享:
紫花苜蓿是世界上最早培育和驯化的优良豆科牧草作物,被誉为“牧草之王”。苜蓿硬实非常普遍,一般表现为种皮不透水而使种子发芽失败,被认为是典型的物理休眠(PY)。由于紫花苜蓿硬实种子和非硬实种子在外观形态上没有明显的差异,传统检测硬实种子最方便有效的方法是进行种子吸水萌发,但这种方法会破坏非硬实种子的种皮结构,且耗时较长,从而导致苜蓿硬实种子和非硬实种子的分子研究受到阻碍。
近日,中国农业大学草业科学与技术学院草种子生理与生产实验室在The Crop Journal上发表了题为“Multiple omics datasets reveal significant physical and physiological dormancy in alfalfa hard seeds identified by multispectral imaging analysis”的研究性论文,作者以紫花苜蓿种子为材料,利用VideometerLab 4多光谱成像技术结合多元分析模型,实现了硬实种子和非硬实种子快速无损鉴别,并在此基础上开展了生理、转录组、代谢组和DNA甲基化等分析,解析出苜蓿硬实种子的多组学差异,提出了苜蓿硬实种子“物理休眠+生理休眠”的综合休眠模式(PY+PD),为深入研究紫花苜蓿硬实种子提供了技术和理论依据。

研究者利用多光谱技术成功实现了紫花苜蓿硬实种子与非硬实种子快速无损鉴别,准确率高达96.8%~99.0%,通过nCDA模型的二次验证,准确率可达100%(表1、图1)。非靶向代谢组学分析发现,差异代谢物主要与类黄酮、脂类和激素的生物合成有关,特别是硬实种子的脱落酸(ABA)含量显著升高(图2)。转录组分析发现差异表达基因富集到“脱落酸反应”途径,进一步支持了植物激素(特别是ABA)对硬实种子的影响(图3)。通过对硬实种子和非硬实种子DNA甲基化分析发现,存在54,899个CpG背景下的差异甲基化区域(DMR),其中344个差异表达基因可能受DNA甲基化的调控(图4)。验证试验表明,紫花苜蓿种子硬实率为8%,但有24.5%的硬实种子擦破种皮后仍不能萌发,将其定义为“非物理休眠的硬实种子”。与硬实种子相比,“非物理休眠的硬实种子”中ABA/IAA和ABA/JA含量显著升高(图5)。综上,研究者提出紫花苜蓿硬实种子可能不是单一的物理休眠,而是“物理休眠+生理休眠”的综合模式。该研究结果为紫花苜蓿种子休眠模式的深入研究奠定了基础。表1 基于SVM、LDA、RF和ELM模型的硬实种子和非硬实种子预测
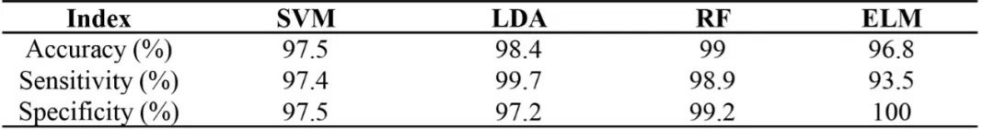
SVM,support vector machine; LDA, linear discriminant analysis; RF, random forest; ELM, extreme learning machine.
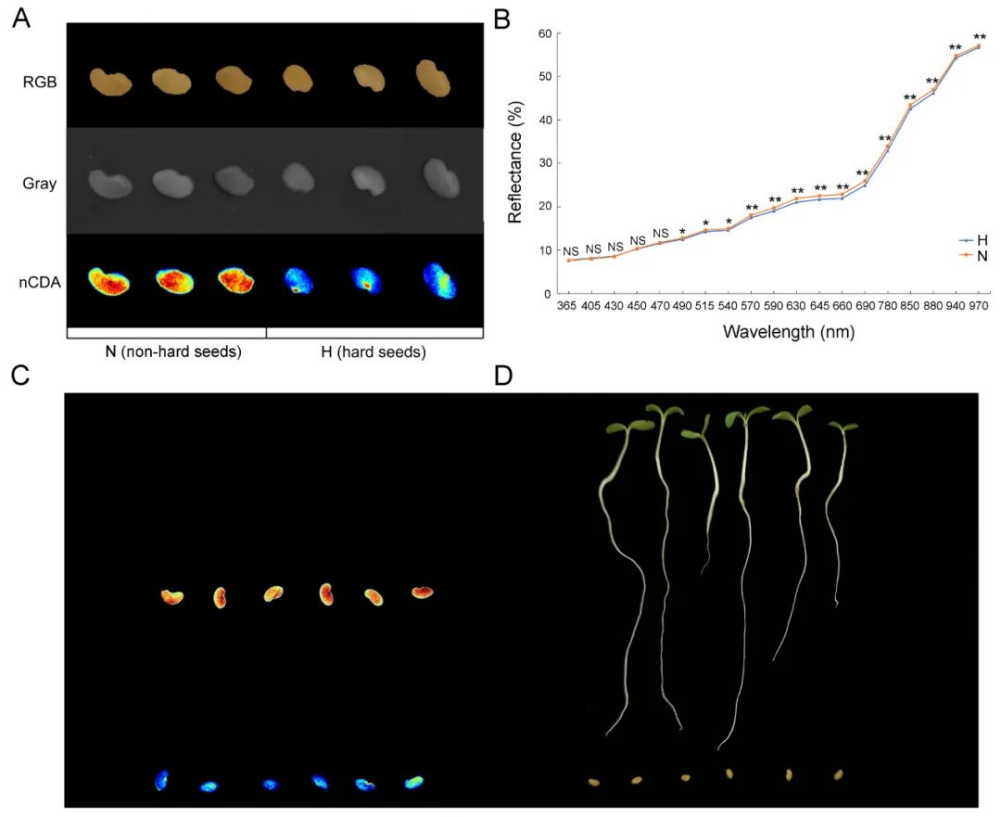
图1多光谱鉴定硬实种子和非硬实种子
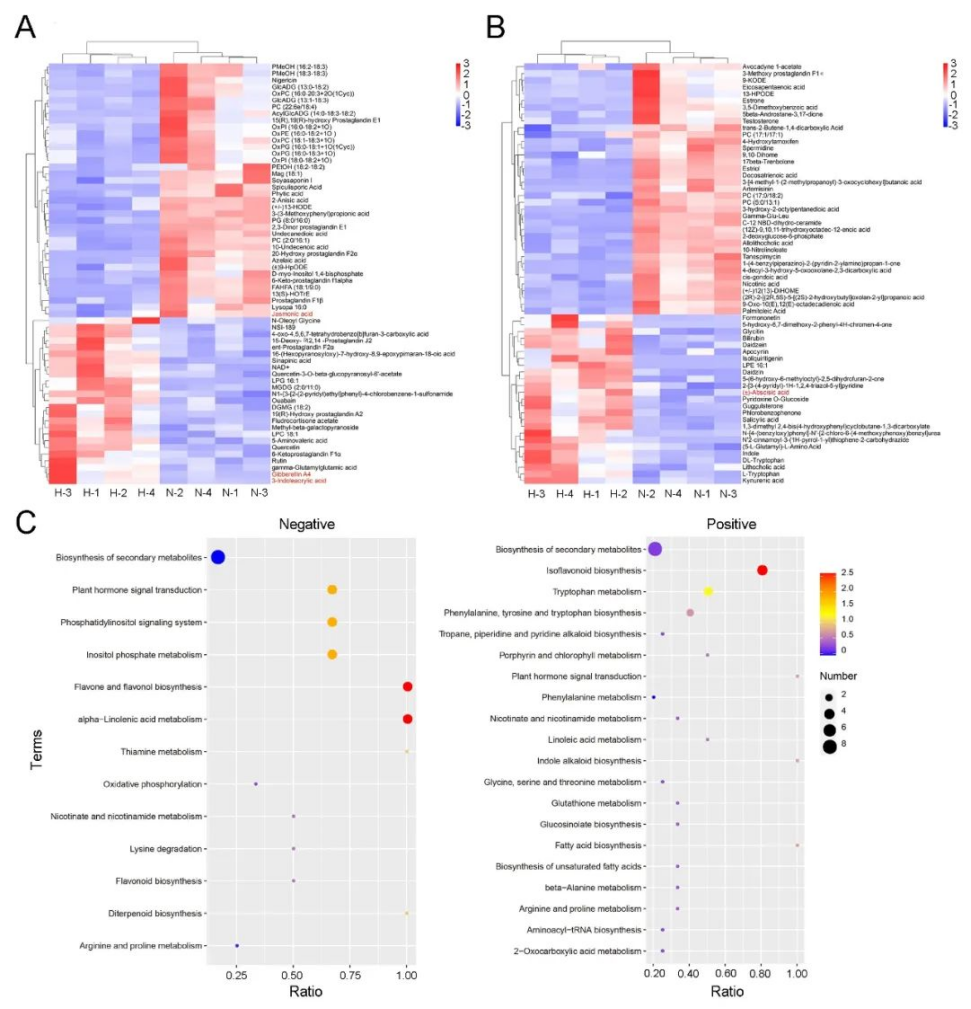
图2硬实种子和非硬实种子的差异代谢物和KEGG富集分析
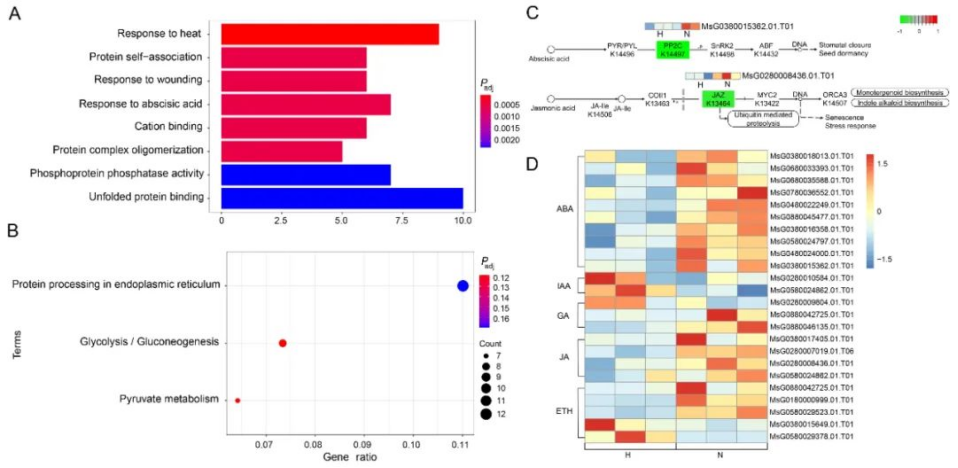
图3硬实种子和非硬实种子的转录组分析
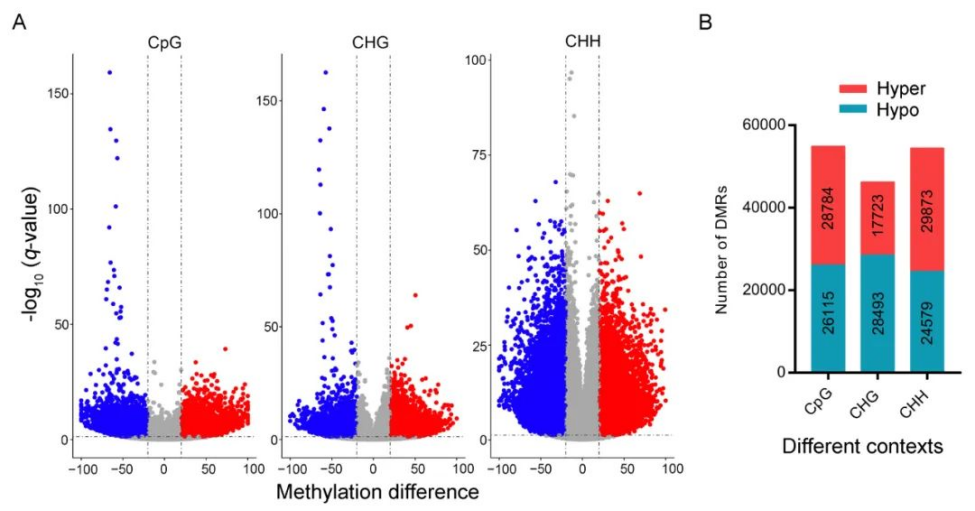
图4 硬实种子和非硬实种子的高甲基化和低甲基化比较
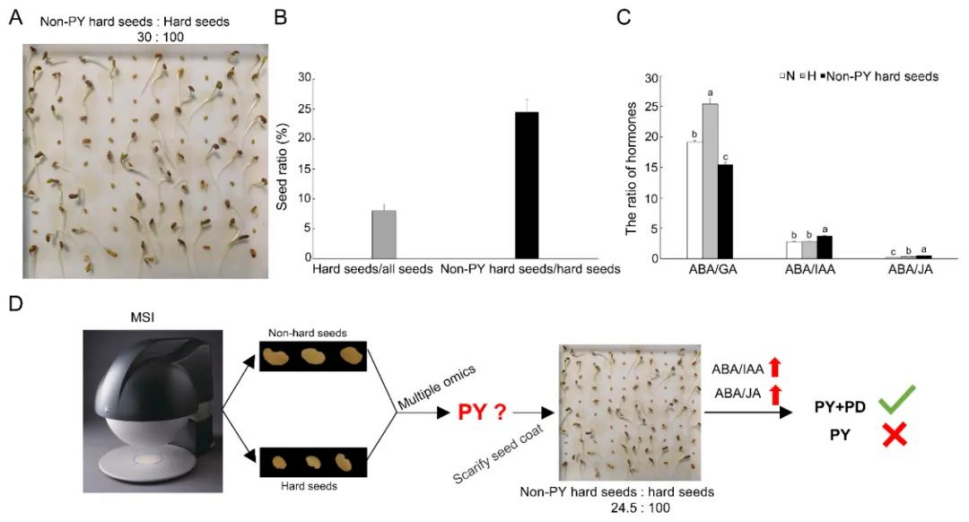
图5 “非物理休眠硬实种子”激素变化
Multiple omics datasets reveal significant physical and physiological dormancy in alfalfa hard seeds identified by multispectral imaging analysis
Abstract
Physical dormancy (PY) commonly present in the seeds of higher plants is believed to be responsible for the germination failure by impermeable seed coat in hard seeds of legume species, instead of physiological dormancy (PD). In this study, a non-destructive approach involving multispectral imaging was used to successfully identify hard seeds from non-hard seeds in Medicago sativa, with accuracy as high as 96.8%–99.0%. We further adopted multiple-omics strategies to investigate the differences of physiology, metabolomics, methylomics, and transcriptomics in alfalfa hard seeds, with non-hard seeds as control. The hard seeds showed dramatically increased antioxidants and 125 metabolites of significant differences in non-targeted metabolomics analysis, which are enriched in the biosynthesis pathways of flavonoids, lipids and hormones, especially with significantly higher ABA, a hormone known to induce dormancy. In our transcriptomics results, the enrichment pathway of “response to abscisic acid” of differential expressed genes (DEG) supported the key role of ABA in metabolomics results. The methylome analysis identified 54,889, 46,216 and 54,452 differential methylation regions for contexts of CpG, CHG and CHH, and 344 DEGs might be regulated by hypermethylation and hypomethylation of promoter and exon regions, including four ABA- and JA-responsive genes. Among 8% hard seeds in seed lots, 24.5% still did not germinate after scarifying seed coat, and were named as non-PY hard seeds. Compared to hard seeds, significantly higher contents of ABA/IAA and ABA/JA were identified in non-PY hard seeds, which indicated the potential presence of PD. In summary, the significantly changed metabolites, gene expressions, and methylations all suggested involvement of ABA responses in hard seeds, and germination failure of alfalfa hard seeds was caused by combinational dormancy (PY+PD), rather than PY alone.
Keywords
Hard seed Multispectral imaging Transcriptomics Metabolomics ABA