品质至上,客户至上,您的满意就是我们的目标
技术文章
当前位置: 首页 > 技术文章
科学家利用WIWAM XY植物表型成像系统在PNAS发表文章
发表时间:2020-05-07 11:24:05点击:1098
来源:北京博普特科技有限公司
分享:
较近,来自比利时、德国、意大利、西班牙的先进科学家联合团队的科研人员利用WIWAM
XY植物表型成像系统发表了题为Histone 2B monoubiquitination complex integrates transcript
elongation with RNA processing at circadian clock and flowering
regulators的文章,发表在生物学先进期刊PNAS上。利用WIWAM XY较早进行拟南芥研究的文章发表在Nature
Biotechnology上,影响因子高达40。科学家利用WIWAM系统陆续发表了系列高影响力、高品质的文章,这为如何利用高通量植物表型成像设备进行植物表型研究提供了很好的参考。利用该系统的段落参见下文。
The small but significant delay in flowering time in spen3-1, the normal flowering time in khd1-1, and the earlier flowering in hub1-4 were also shown in an auto- mated weighing, imaging, and watering phenotyping platform (WIWAM XY; https://www.wiwam.be) (Fig. 2E). The over- expression lines of HUB1, SPEN3, and KHD1 included in this experiment had normal flowering times (Fig. 2E). The number of leaves at bolting, which is tightly correlated with flowering time (41, 42), was lower in the early-flowering genotypes and slightly higher in spen3-1 and spen3-3 than that in the wild type, as expected (SI Appendix, Fig. S4 E–G).
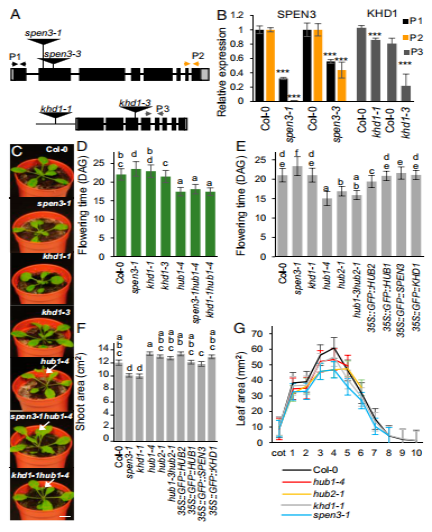
Fig. 2.Phenotypes of single and double mutants and of overexpression lines. (A) Schematic view of the SPEN3 and KHD1 T-DNA insertion lines, spen3-1
(SALK_025388), spen3-3 (GABI_626H01), khd1-1 (SALK_046957), and khd1-3 (SAIL_1285_H03C1). (B) Relative expression of KHD1 and SPEN3 in Col-0 and mutant lines estimated with the qPCR assay in six biological replicates. Asterisks indicate statistically significant differences by Student’s t test (***P < 0.001). (C) Representative seedlings at 19 DAG grown in jiffy containers. Arrows mark early-emerging inflorescences. (Scale bar, 1 cm.) (D) Flowering time of lines germinated in jiffy pots (n = 51). (E and F) Flowering time and projected rosette area of seed lings at 23 DAS (n = 24) respectively, grown on the WIWAM phenotyping platform. Error bars represent SEs. Ordinary one-way ANOVA with 95% confidence shows a significant difference between the genotypes, represented by letters (D–F). (G) Individual leaf areas of seedlings at 21 DAS of mutant lines, in vitro grown on the IGIS platform.
作为先进家将大规模自动化理念和工业级零件和设备整合入植物成像系统的厂家,SMO公司在植物表型成像分析领域处于先进的技术前列,大面积叶绿素荧光成像系统使WIWAM成为植物表型分析与功能成像领域较为先进的仪器设备,植物生长、胁迫响应等测量参数达几百个。工业级部件品质使系统非常耐用,基本免维护,与同类产品相比,特点突出。目前WIWAM植物表型平台分为WIWAM XY,WIWAM Line、WIWAM Conveyor、WIWAM Imaging Box以及WIWAM FIELD等几个系列,涵盖从室内表型成像到室外表型成像各个领域,多维度、多类型传感器和设备的应用为植物表型科研提供了很好的借鉴思路。
WIWAM XY是一款高通量可重复性表型机器人, 用于对小型植物, 如拟南芥植物的研究。该机器人可定期对多种植物参数进行自动化灌溉和并测量多种植物生长参数。WIWAM XY代替了很多手工处理,省时省钱,精度较高。

WIWAM植物表型成像系统叶绿素荧光成像版
北京博普特科技有限公司是比利时WIWAM植物表型成像系统中国区总代理,全面负责其系列产品在中国市场的推广、销售和售后服务。
Woloszynska
M, Gall SL, Neyt P, Boccardi TM, Grasser M, Längst G, Aesaert S, Coussens G,
Dhondt S, Slijke EVD, et al (2019) Histone 2B monoubiquitination complex
integrates transcript elongation with RNA processing at circadian clock and
flowering regulators. PNAS 116: 8060–8069
Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators
View ORCID ProfileMagdalena Woloszynska, View ORCID ProfileSabine Le Gall, View ORCID ProfilePia Neyt, Tommaso M. Boccardi, Marion Grasser, View ORCID ProfileGernot Längst, View ORCID ProfileStijn Aesaert, Griet Coussens, View ORCID ProfileStijn Dhondt, View ORCID ProfileEveline Van De Slijke, View ORCID ProfileLeonardo Bruno, View ORCID ProfileJorge Fung-Uceda, View ORCID ProfilePaloma Mas, View ORCID ProfileMarc Van Montagu, View ORCID ProfileDirk Inzé, View ORCID ProfileKristiina Himanen, View ORCID ProfileGeert De Jaeger, Klaus D. Grasser, and View ORCID ProfileMieke Van Lijsebettens
PNAS April 16, 2019 116 (16) 8060-8069; first published March 28, 2019 https://doi.org/10.1073/pnas.1806541116
Significance
In eukaryotes, the genomic DNA is organized in chromatin that consists of nucleosomal units formed by histone proteins. The accessibility of genes for RNA polymerase II transcription by the dynamic modification of histone tails during the transcript elongation phase is emerging as an important regulatory mechanism. Here, we show that RNA-binding proteins, together with the conserved HISTONE MONOUBIQUITINATION1–HISTONE MONOUBIQUITINATION2 complex that mediates histone H2B monoubiquitination at the circadian clock, and flowering time-regulatory genes during transcript elongation are required for the processing of their pre-mRNA and antisense RNA, respectively.
Abstract
HISTONE MONOUBIQUITINATION1 (HUB1) and its paralog HUB2 act in a conserved heterotetrameric complex in the chromatin-mediated transcriptional modulation of developmental programs, such as flowering time, dormancy, and the circadian clock. The KHD1 and SPEN3 proteins were identified as interactors of the HUB1 and HUB2 proteins with in vitro RNA-binding activity. Mutants in SPEN3 and KHD1 had reduced rosette and leaf areas. Strikingly, in spen3 mutants, the flowering time was slightly, but significantly, delayed, as opposed to the early flowering time in the hub1-4 mutant. The mutant phenotypes in biomass and flowering time suggested a deregulation of their respective regulatory genes CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and FLOWERING LOCUS C (FLC) that are known targets of the HUB1-mediated histone H2B monoubiquitination (H2Bub). Indeed, in the spen3-1 and hub1-4 mutants, the circadian clock period was shortened as observed by luciferase reporter assays, the levels of the CCA1α and CCA1β splice forms were altered, and the CCA1 expression and H2Bub levels were reduced. In the spen3-1 mutant, the delay in flowering time was correlated with an enhanced FLCexpression, possibly due to an increased distal versus proximal ratio of its antisense COOLAIR transcript. Together with transcriptomic and double-mutant analyses, our data revealed that the HUB1 interaction with SPEN3 links H2Bub during transcript elongation with pre-mRNA processing at CCA1. Furthermore, the presence of an intact HUB1 at the FLC is required for SPEN3 function in the formation of the FLC-derived antisense COOLAIR transcripts.